SEQUiT! in brief
SEQUiT 4.1 - de novo peptide sequencing software is ...
... precise
|
|
in computation of complete peptide sequences |
|
|
in successful sequence computing from tandem mass spectrometry data using even incomplete b- or y-fragment series |
|
|
in accurate sequencing of tryptic and nontryptic (e.g. MHC peptides) peptides |
|
|
in sequencing of chemically or post-translationally modified peptides |
|
|
in different scoring systems for ESI- and MALDI-MS techniques (already successfully tested with data provided by MALDI-QTOF, MALDI-TOF (Post Source Decay), MALDI-TOF/ TOF (Laser Induced Dissociation, Collision Induced Dissociation), ESI-TRAP, ESI-QTOF and ESI-FTICR) |
... highly customizable
|
|
in extending individual amino acid list |
|
|
in extendending own list of modifications |
|
|
in using BLAST search engine in combination with different databases and taxonomy restrictions by GI (GenBank identifier) lists |
... highly automated
|
|
SEQUiT! supports common MS/MS data formats (e.g. *.sqi, *.mgf, *.dta, *.pkl) |
|
|
You can process batch data: de novo sequencing with subsequent database searches |
|
|
SEQUiT! generates report files with batch analysis results |
... simple
SEQUiT 4.1 allows intuitionally and uncomplicated usage.
Examples: Screenshots
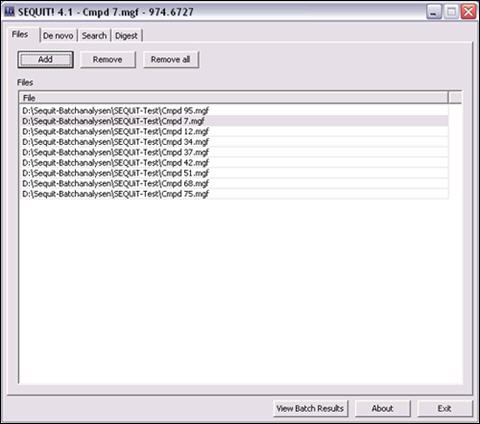
File Manager
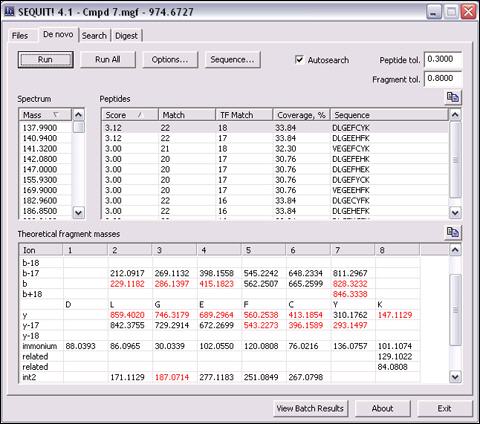
De novo sequencing module
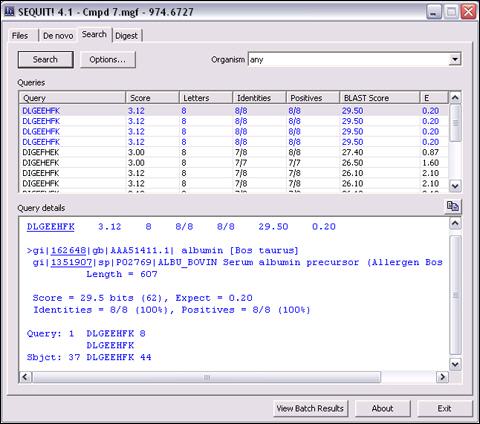
Database Search (BLAST)
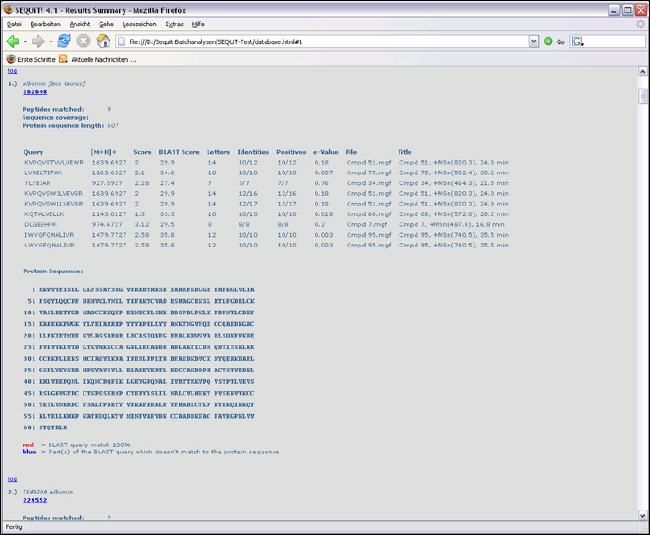
Result Overview
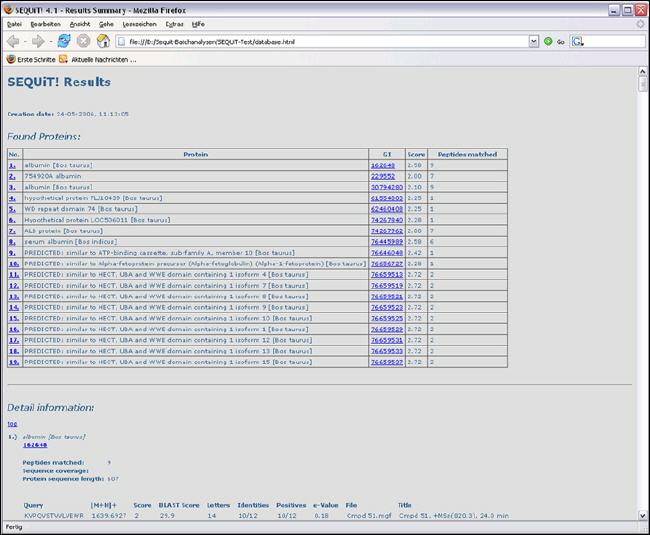
Results in Detail
References
SEQUiT! software
Demine R., Walden P. Sequit: software for de novo peptide sequencing by matrix-assisted laser desorption/ionization post-source decay mass spectrometry. Rapid Commun Mass Spectrom. 2004; 18: 907-913.
Used in
Demine R., Walden P. Testing the role of gp96 as peptide chaperone in antigen processing. J Biol Chem. 2005; 280: 17573-17578.
Hardware/software configuration
Minimal: Pentium 4 CPU 1.70 MHz, 512 MB RAM, Windows 2000 or Windows XP, IE 6.0
Optimal: Pentium 4 CPU 3.40 MHz, 2 GB RAM, Windows 2000 or Windows XP, IE 6.0
Pentium is a registered trademark of Intel Corporation.
Windows 2000 and Windows XP are registered trademarks of Microsoft Corporation
(C) 2006 - Proteome Factory AG - All rights reserved